Abstract
Background: Cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA) analysis has an emerging diagnostic role in multiple malignancies including lymphomas. As levels of these biomarkers have been correlated with clinical prognosis, knowledge of baseline values could have utility given wide clinical variation among lymphoma subtypes. We examined cfDNA and ctDNA levels for a pooled international cohort representing both classical Hodgkin (cHL) and non-Hodgkin (NHL) lymphomas. In addition, we analyzed corresponding clinical laboratory data for confounding factors.
Methods: A total of 196 patients from six centers were analyzed - Stanford University (65), University of Eastern Piedmont (36), the National Cancer Institute (33), Hospital Le Bocage, Dijon (25), MD Anderson Cancer Center (21), and University Hospital Essen (16). Pre-treatment cfDNA concentration was obtained by fluorimetry after DNA extraction. All samples underwent deep sequencing with error correction by CAPP-Seq (Newman et al Nat Biotech 2016). After genotyping, ctDNA levels were calculated as the product of the cfDNA concentration and the mean allelic fraction of somatic single nucleotide mutations. Where available, clinical laboratory data from the time of ctDNA measurement were correlated.
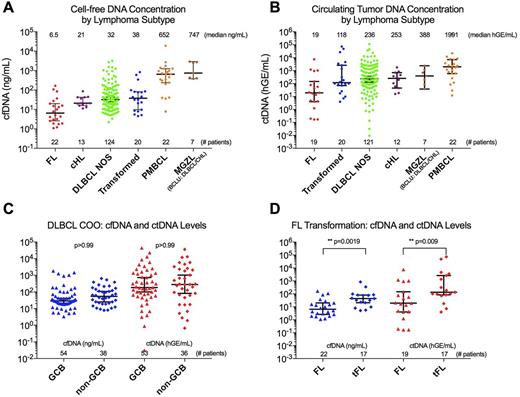
Results: Across lymphoma subtypes, total cfDNA levels were observed to distinguish between aggressive and indolent histologies (Fig 1A). The highest cfDNA levels were seen in a statistically distinct subset of aggressive thoracic tumors including mediastinal grey zone lymphoma (MGZL) and primary mediastinal tumors (PMBCL). These did not differ significantly from each other (p>0.999), but were higher than other aggressive lymphomas including diffuse large B cell lymphoma (DLBCL), cHL, and transformed low grade lymphomas (p<0.013 for all comparisons). Follicular lymphoma (FL) and cHL had the lowest median cfDNA levels.
Circulating tumor DNA was detected in 97.4% of patients (191/196) with no difference in detection rate by subtype. Median ctDNA level followed a similar distribution to cfDNA, with highest levels in aggressive thoracic MGZL and PMBCL and lowest in FL (Fig 1B). Variation in ctDNA levels was notably broader than cfDNA, with DLBCL varying across a 10-million fold concentration range.
To further characterize our DLBCL cohort we separated cases on cell of origin (COO) determined by Hans classification, and by histological transformation status. No difference in cfDNA or ctDNA levels was seen for GCB versus non-GCB histology (Fig 1C), or for de novo versus transformed DLBCL. However, transformed FL had significantly higher levels of both cfDNA and ctDNA than primary FL (Fig 1D, p<0.0001 for cfDNA, p=0.0032 for ctDNA).
We correlated our findings with clinical laboratory data for potential confounders. Although cfDNA is renally cleared, we found no significant correlation between serum creatinine and cfDNA level (p=0.77). While the majority of cfDNA originates in the hematologic compartment, we found no significant correlation with WBC (p=0.10) and a negative correlation with hematocrit (p=0.0016) and serum albumin levels (p=0.0003). Confirming prior observations, LDH was positively correlated with cfDNA level (r=0.51, p=0.0004). All described laboratory correlations remained significant after Bonferroni correction.
Conclusions: Cell free and ctDNA levels vary significantly across lymphoma histologies, suggesting that both total levels and genotype may play a role in future assays. The highest levels were observed in a subset of aggressive thoracic tumors which may reflect higher disease volumes, proliferative rates, and/or proximity to venous circulation. Minimal variation with renal and hepatic functions reinforces the reliability of cfDNA as a biomarker, while correlations with anemia, hypoalbuminemia, and LDH elevation are likely reflective of inflammatory disease burden.
Figure 1: Variation by lymphoma subtype of cfDNA and ctDNA levels.
A) A scatter plot demonstrates variation in cell-free DNA (cfDNA) levels across lymphoma subtypes.
B) Circulating tumor DNA (ctDNA) levels vary across lymphoma subtypes.
C) Neither cfDNA nor ctDNA levels differ significantly between DLBCL cell of origin subtypes.
D) Primary and transformed FL display significant variation in cfDNA and ctDNA levels.
(hGE/mL: Haploid genome equivalents / mL)
Hüttmann: Bristol-Myers Squibb, Takeda, Celgene, Roche: Honoraria; Gilead, Amgen: Other: Travel cost. Casasnovas: BMS: Consultancy, Honoraria; Takeda: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Sanofi: Consultancy, Honoraria; Gilead: Consultancy, Honoraria, Research Funding; Roche: Consultancy, Honoraria, Research Funding. Westin: Novartis Pharmaceuticals Corporation: Membership on an entity's Board of Directors or advisory committees; Apotex: Membership on an entity's Board of Directors or advisory committees; Celgene: Membership on an entity's Board of Directors or advisory committees; Kite Pharma: Membership on an entity's Board of Directors or advisory committees. Gaidano: Janssen: Consultancy, Honoraria; Gilead: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria; Roche: Consultancy, Honoraria; Amgen: Consultancy, Honoraria. Advani: Juno Therapeutics: Consultancy; Bayer Healthcare Pharmaceuticals: Research Funding; Infinity: Research Funding; Gilead: Consultancy; Celgene: Research Funding; Spectrum: Consultancy; Millennium: Research Funding; FortySeven: Research Funding; Kura: Research Funding; Genentech: Research Funding; Cell Medica: Research Funding; Janssen: Research Funding; Seattle Genetics: Research Funding; Nanostring: Consultancy; Regeneron: Research Funding; Bristol-Myers Squibb: Consultancy, Research Funding; Pharmacyclics: Consultancy; Merck: Research Funding; Agensys: Research Funding; Sutro: Consultancy; Pharmacyclics: Research Funding. Diehn: Roche: Consultancy; Novartis: Consultancy; Quanticel Pharmaceuticals: Consultancy; Varian Medical Systems: Research Funding. Alizadeh: CiberMed: Consultancy; Genentech: Consultancy; Celgene: Consultancy; Roche: Consultancy; Gilead: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.